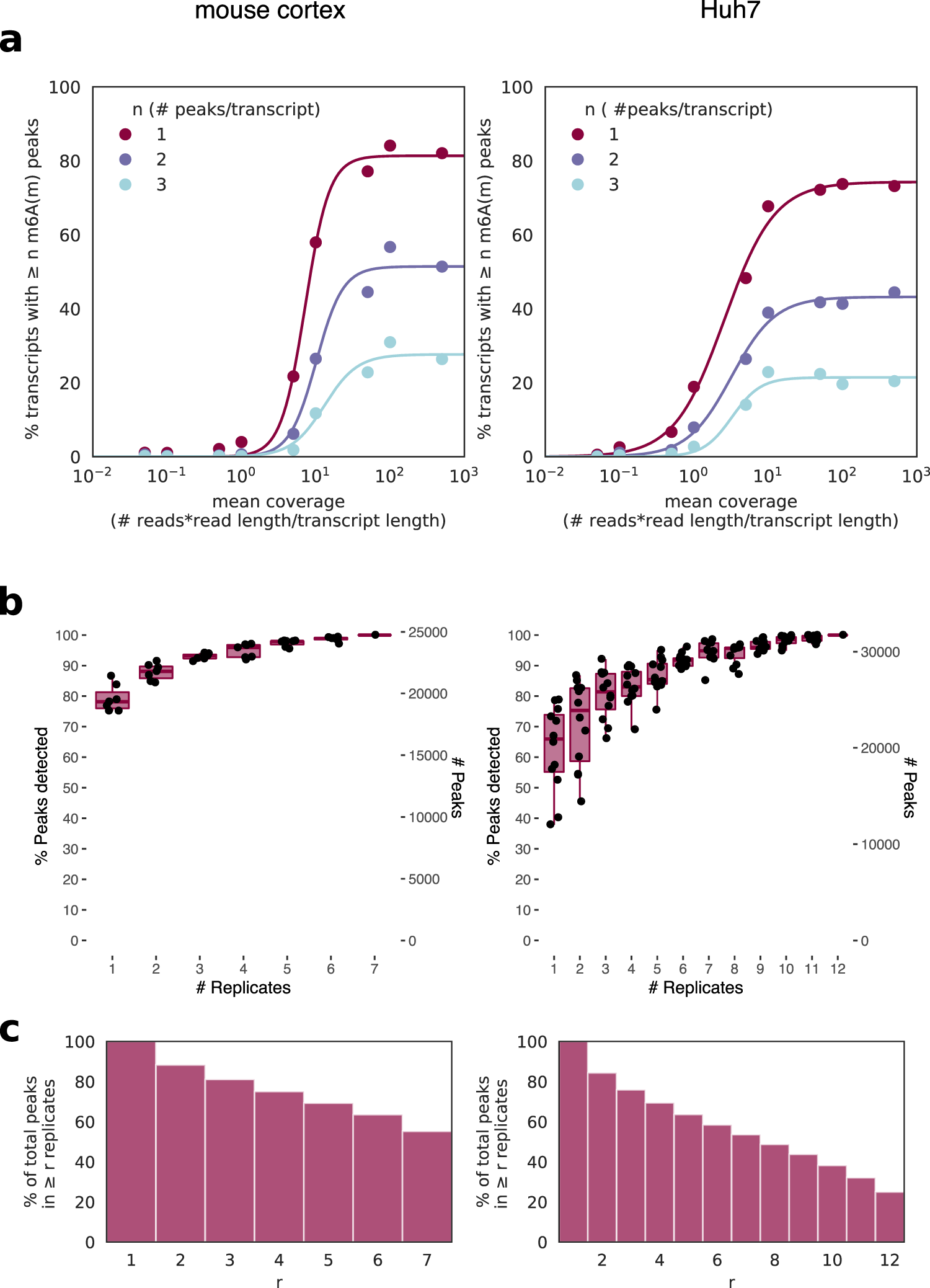
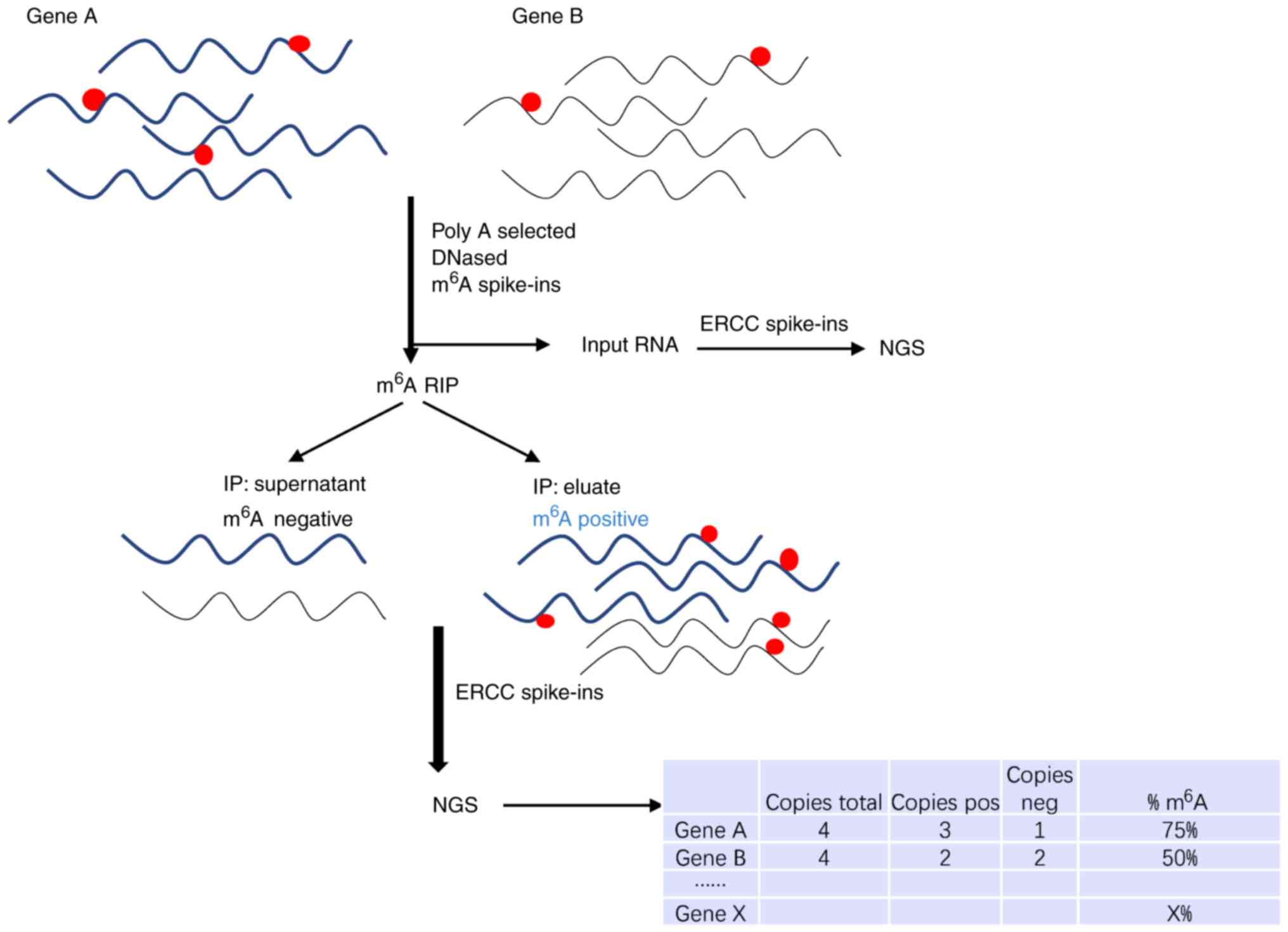
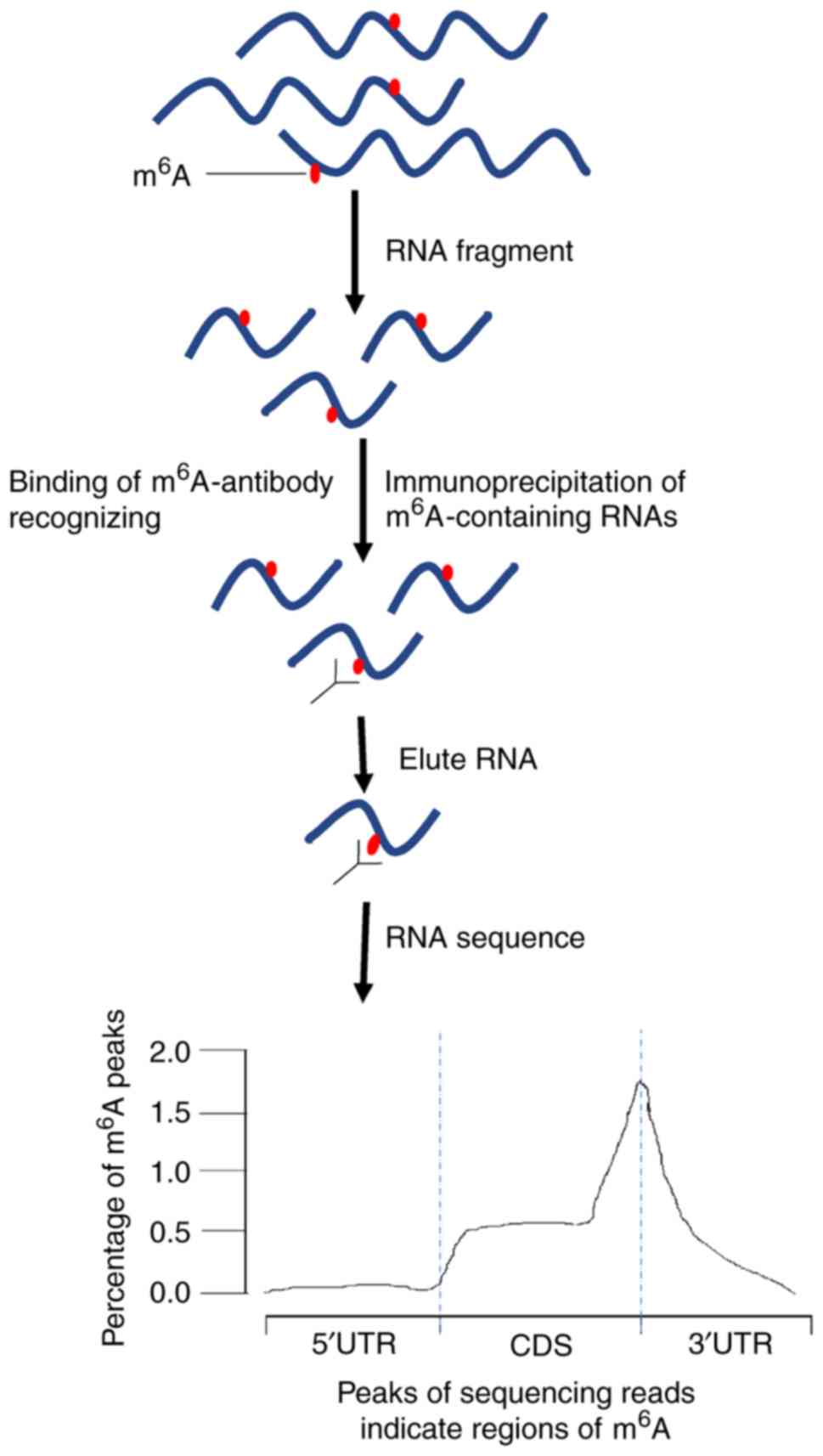
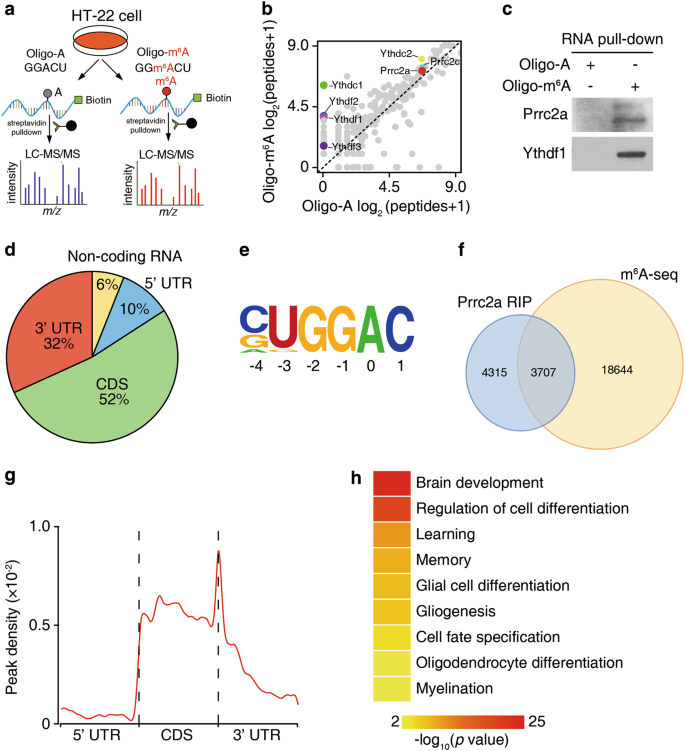
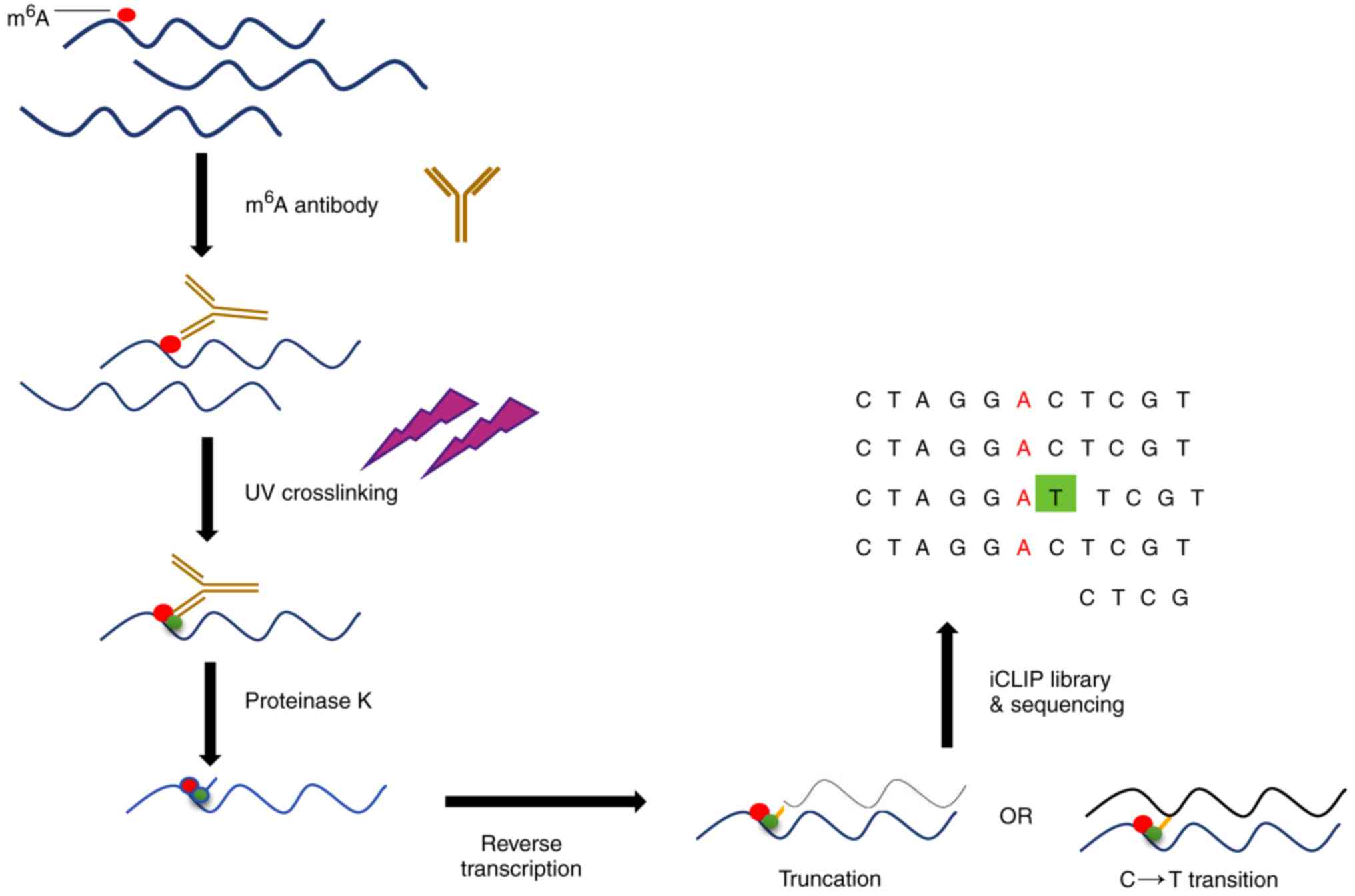
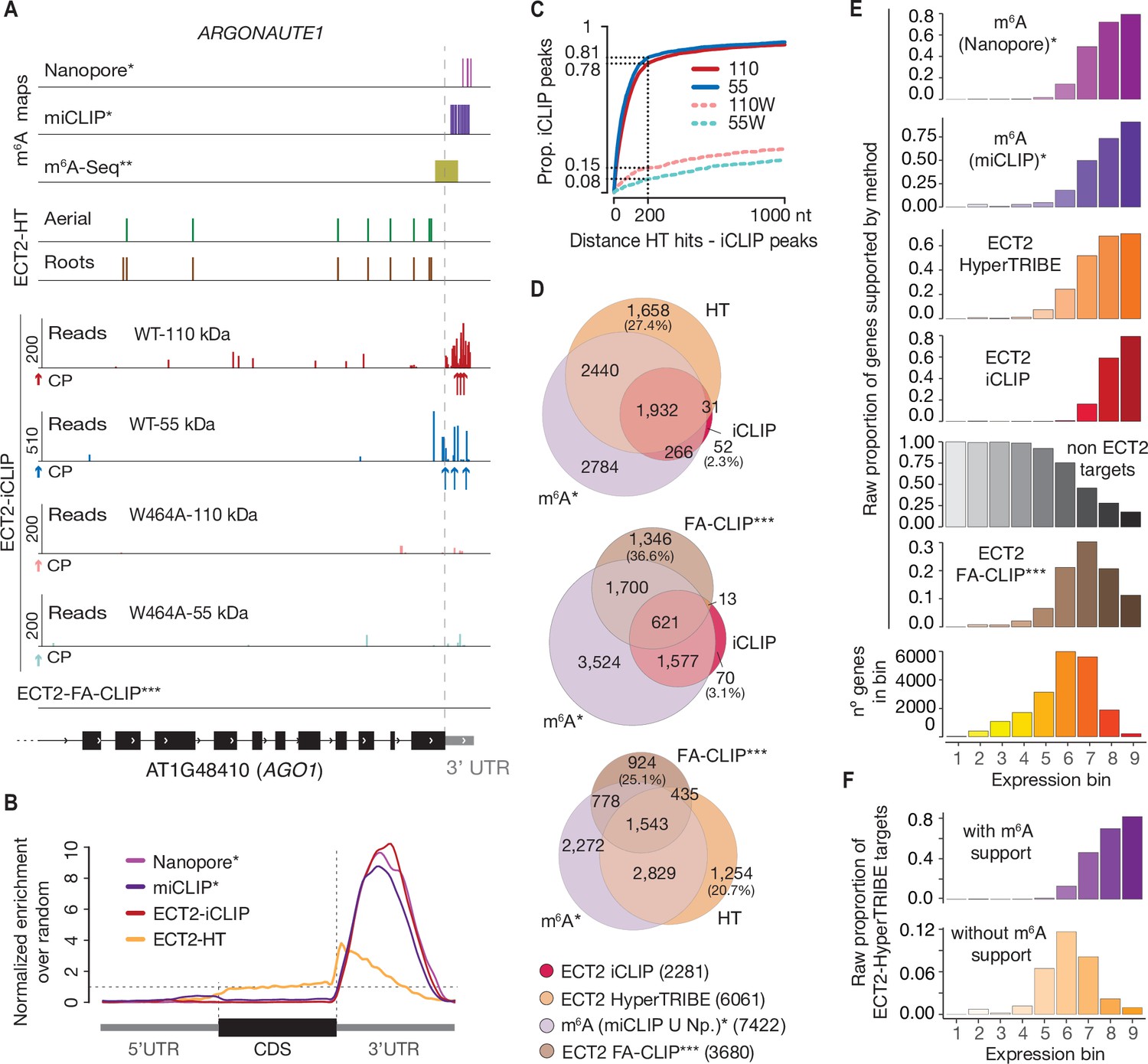
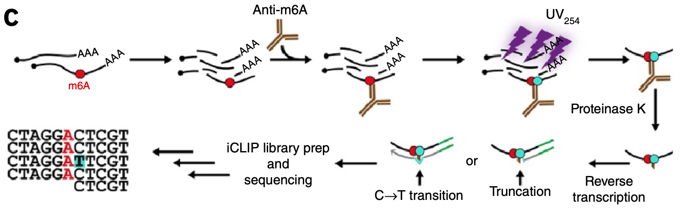
Evolutionary conservation of the DRACH signatures of potential N6-methyladenosine (m6A) sites among influenza A viruses | Scientific Reports
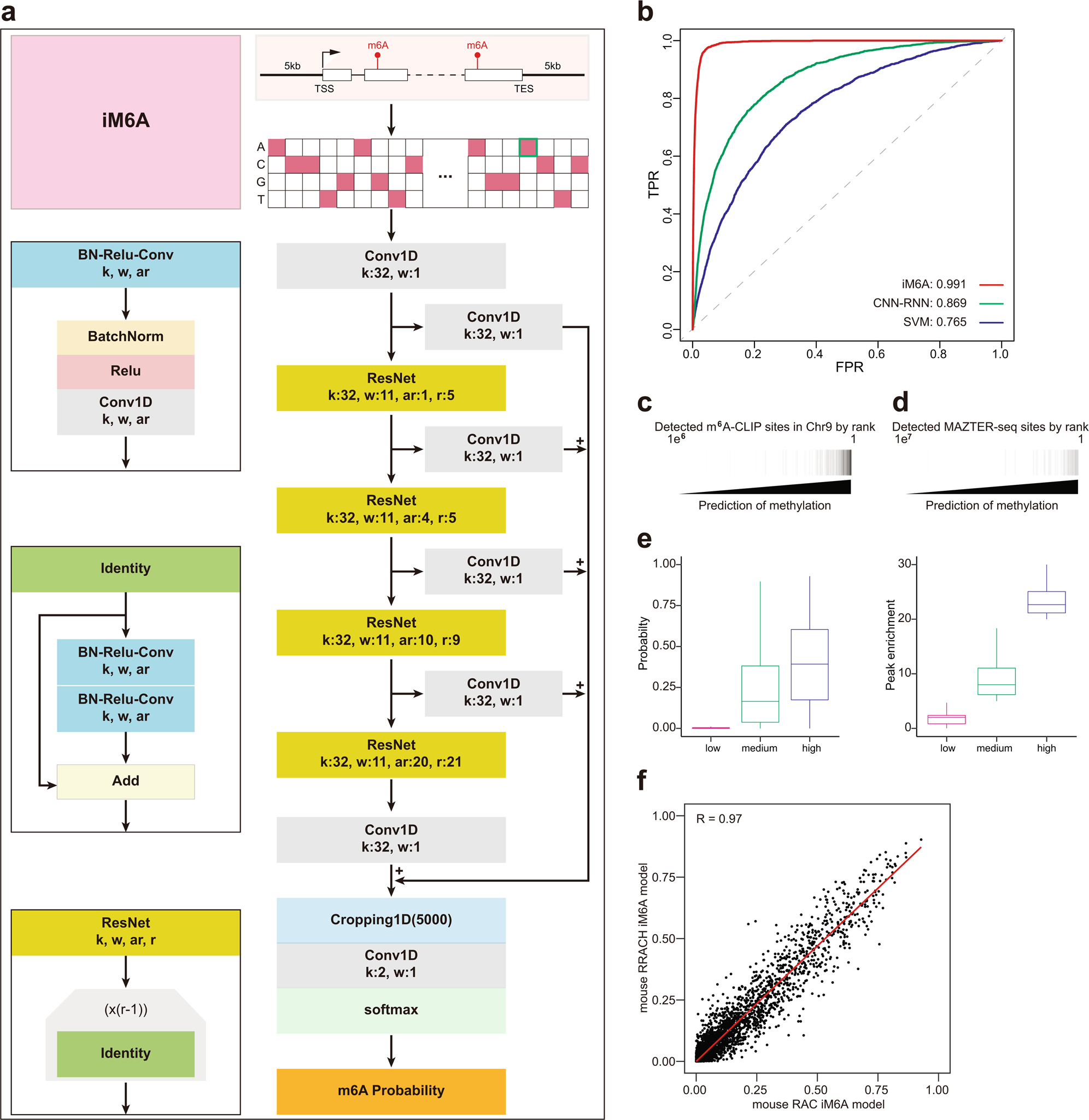
Deep learning modeling m6A deposition reveals the importance of downstream cis-element sequences | Nature Communications

METTL3 and WTAP bind m6A consensus sequence in mRNA and affect gene... | Download Scientific Diagram
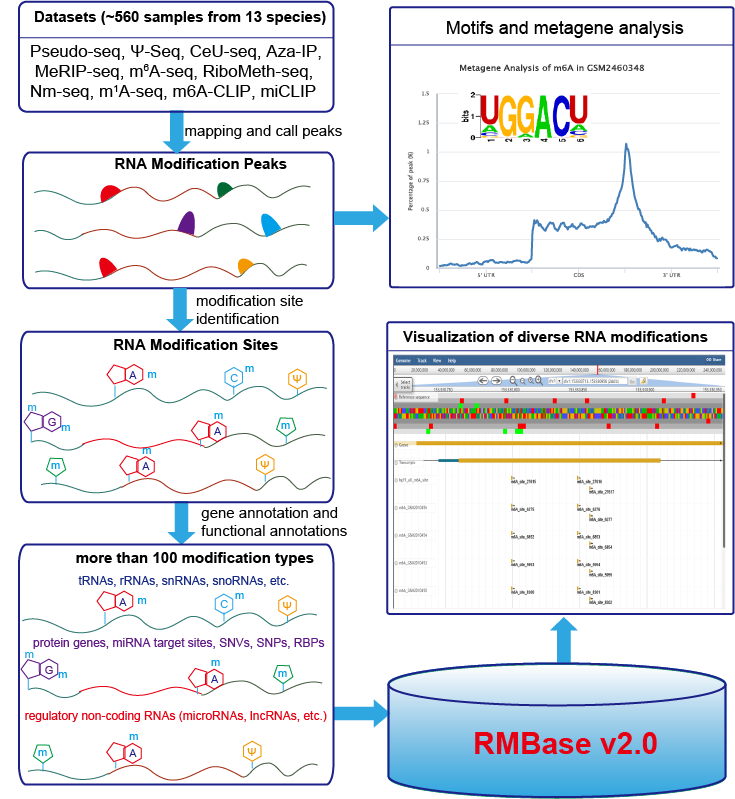
RNA Modification Database, RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data (Pseudo-seq, Ψ-seq, CeU-seq, Aza-IP, MeRIP-seq, m6A-seq, RiboMeth-seq, m1A-seq).

Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine | Nature Chemical Biology

High‐Resolution N6‐Methyladenosine (m6A) Map Using Photo‐Crosslinking‐Assisted m6A Sequencing - Chen - 2015 - Angewandte Chemie International Edition - Wiley Online Library