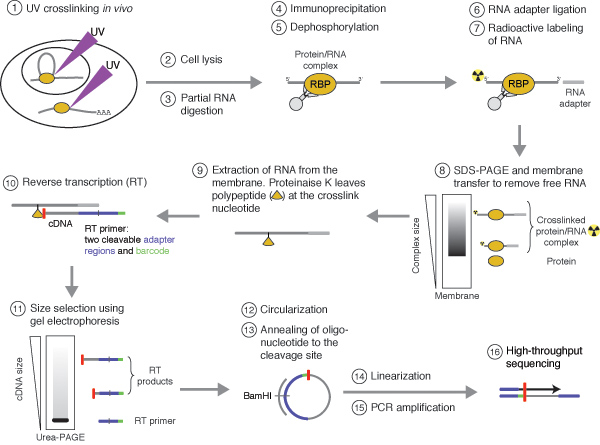
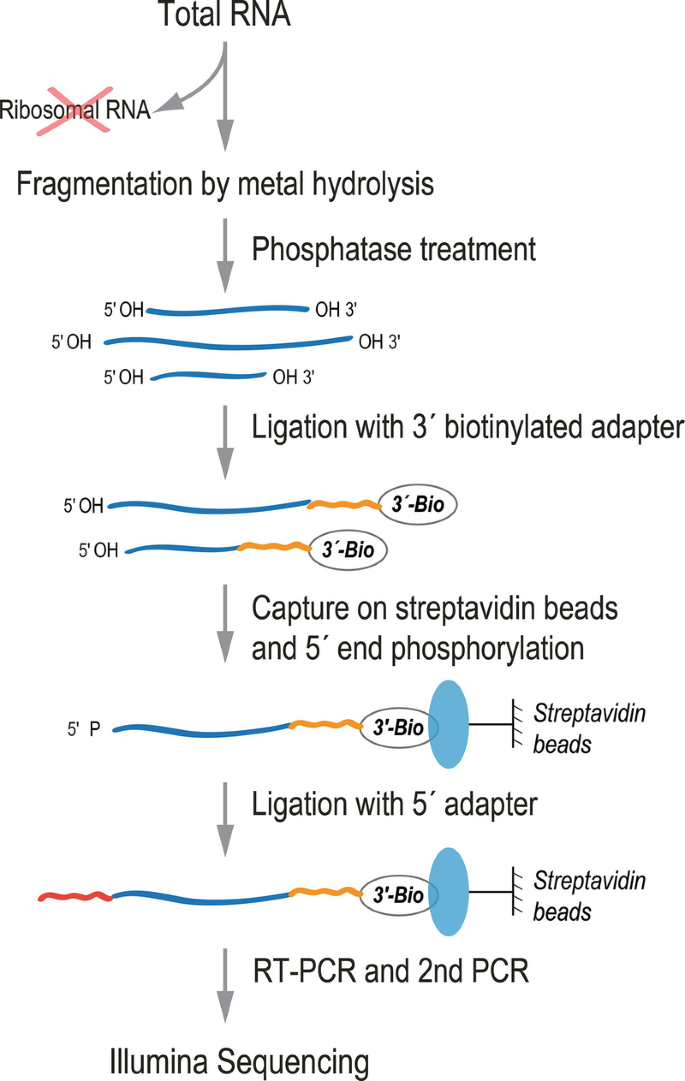
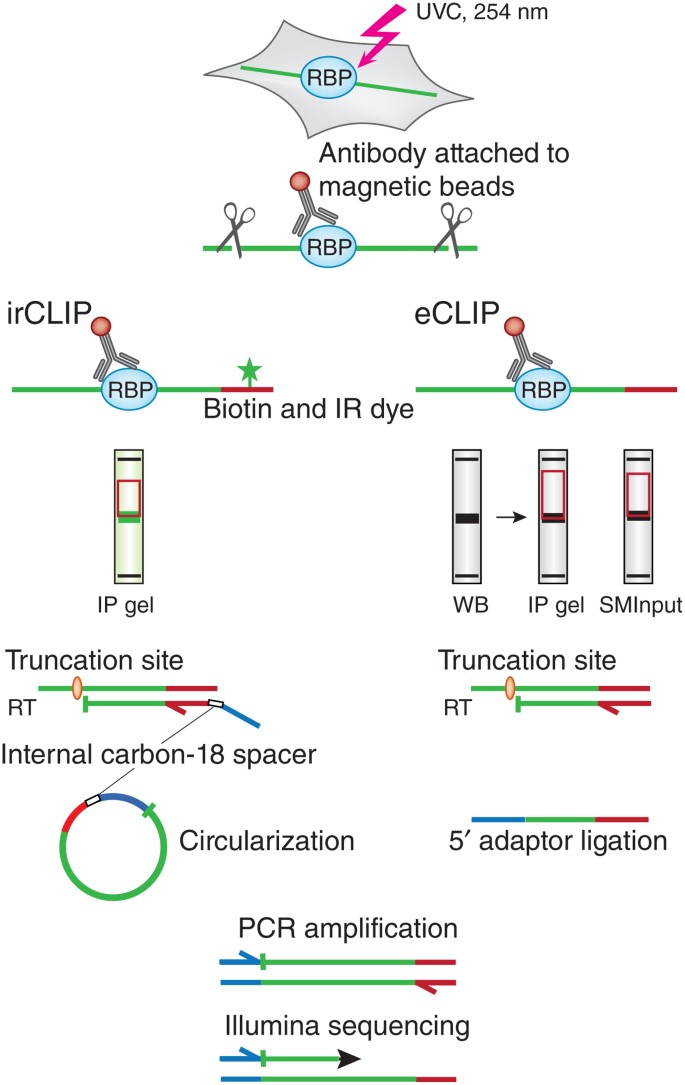
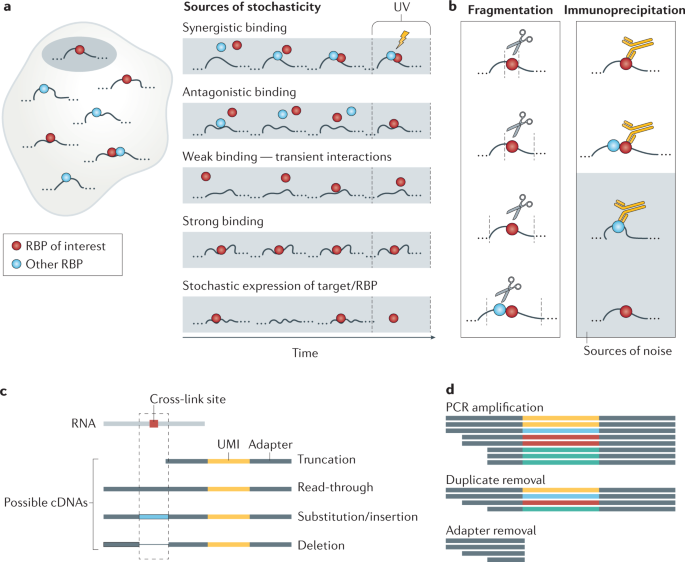
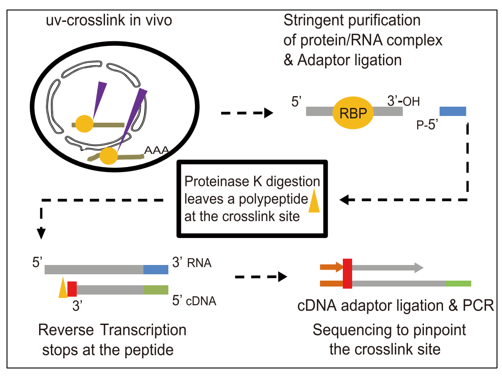
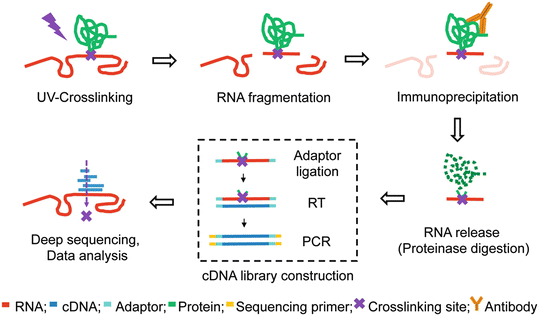
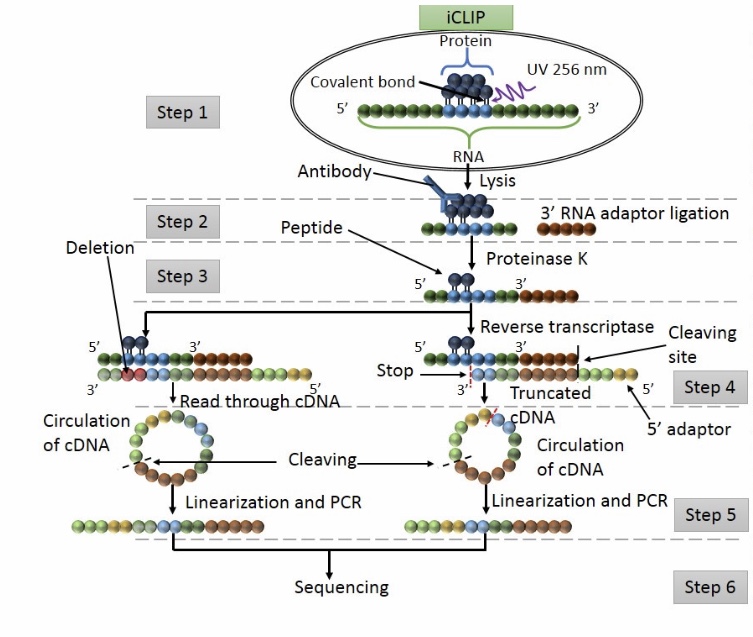
tRIP‐seq reveals repression of premature polyadenylation by co‐transcriptional FUS‐U1 snRNP assembly | EMBO reports

ACE-score-based Analysis of Temporal miRNA Targetomes During Human Cytomegalovirus Infection Using AGO-CLIP-seq
Enhanced CLIP Uncovers IMP Protein-RNA Targets in Human Pluripotent Stem Cells Important for Cell Adhesion and Survival
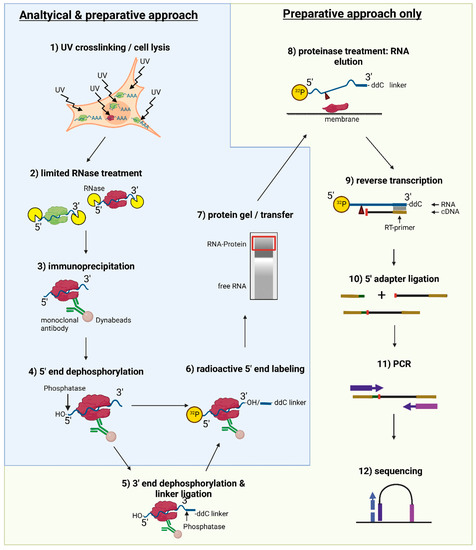
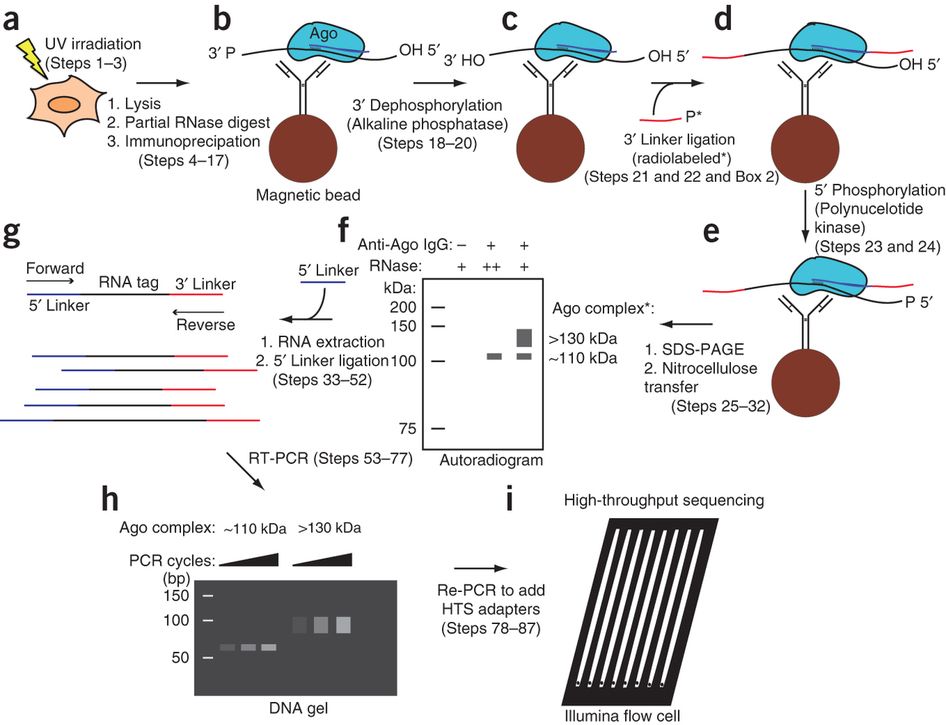
MPs | Free Full-Text | Studying miRNA–mRNA Interactions: An Optimized CLIP-Protocol for Endogenous Ago2-Protein

Schematic workflow of optimized CLIP. Cells are irradiated with 254 nm... | Download Scientific Diagram
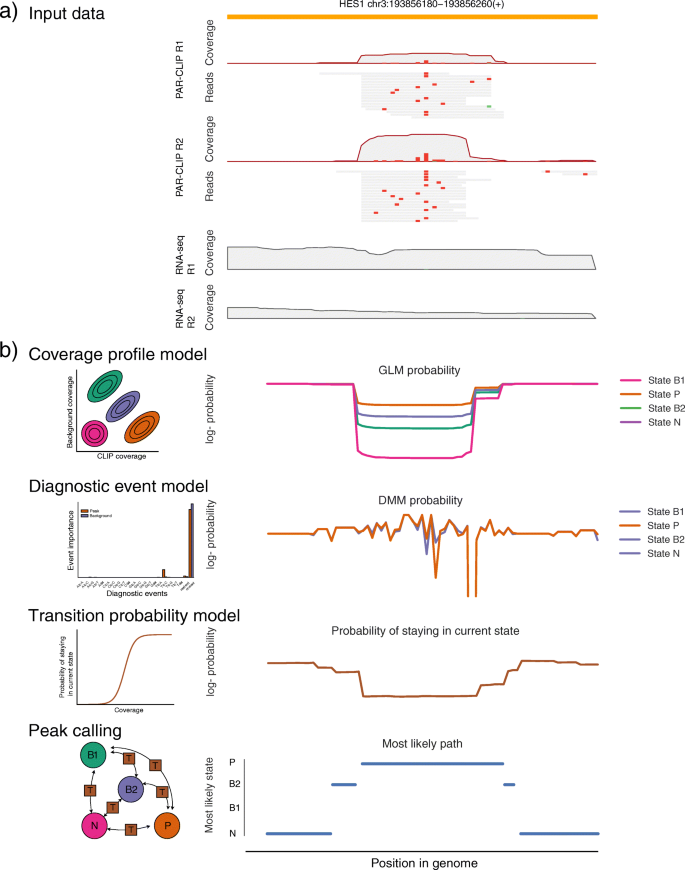
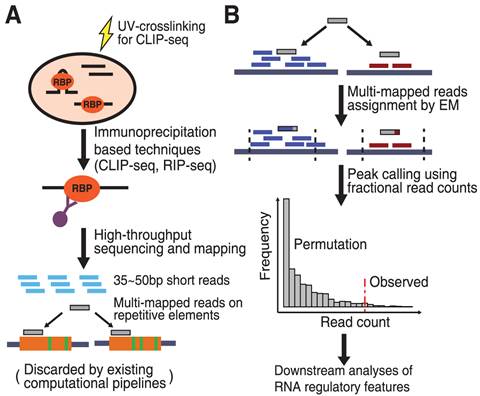
omniCLIP: probabilistic identification of protein-RNA interactions from CLIP -seq data | Genome Biology | Full Text

PAR-CLIP and streamlined small RNA cDNA library preparation protocol for the identification of RNA binding protein target sites - ScienceDirect
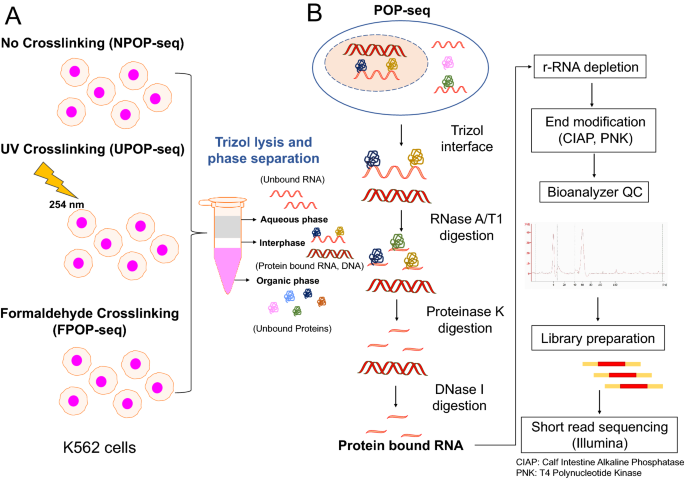
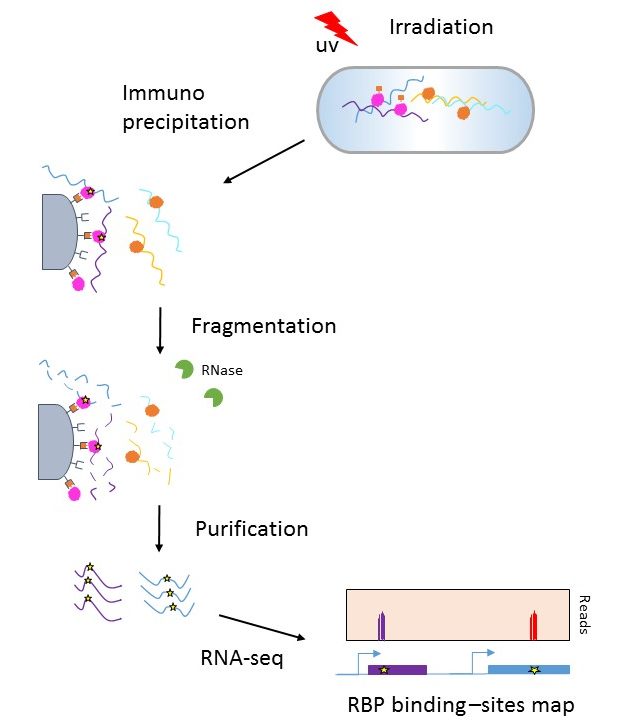
Transcriptome-wide high-throughput mapping of protein–RNA occupancy profiles using POP-seq | Scientific Reports